Meet EDDI
EDDI is our Epigenetic-Digital-Data-Interface
an ML model capable of forward and backward prediction of cell culture parameters and quality attributes.

Digital Analytics Validated
Confirming, the digital analysis of process parameters and through epigenetic data (specifically methylation patterns) has been successfully established, enabling enhanced monitoring and optimisation of production processes.

Predictive Modelling Substantiated
We have demonstrated that predicting bioprocess outcomes from methylation data at a single time is not only feasible but also reduces development timelines effectively. This validation of our approach in real-world applications holds immense potential for improving the efficiency of biopharmaceutical production.

Pathway Insights Achieved
Our research has deepened the understanding of biochemical pathways, confirming that pathway-driven decision-making based on epigenetic data can significantly improve the accuracy and efficiency of biopharmaceutical production.
Follow our journey in developing predictive models and digital twins for CHO, HEK and other human cell lines
Please subscribe to our EDDI newsletter
Some insights
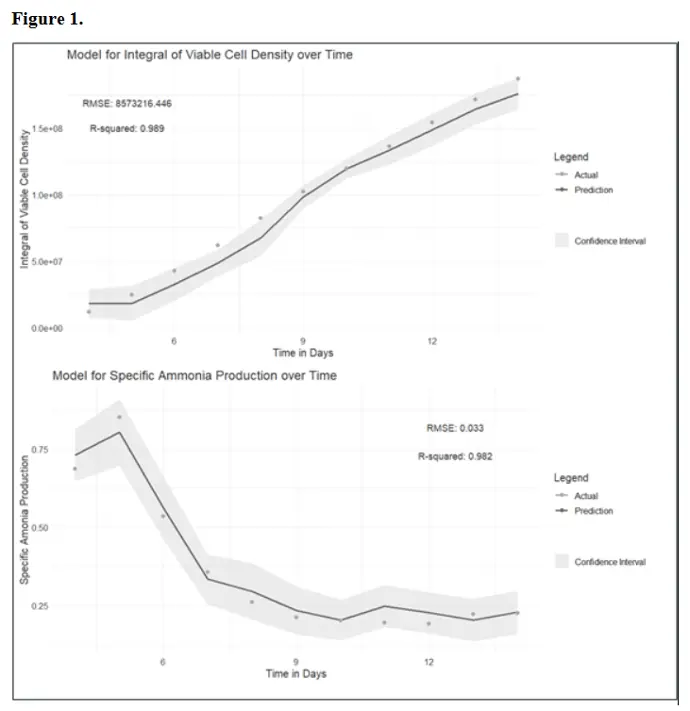
By analyzing about 50,000 methylation sites within the CHO genome, and up to 200.000 sites for the human genom we can build experiment-specific predictions (R2 > 0.9) based on as little as 10 historical datasets. These predictions include cell growth and waste production at the end of a 14-day bioprocess with a single sampling on day 4 (Figure 1) or reconstructing the conditions of a particular bioprocess, such as the media used (Figure 2). Because the methylation profiles and specific models are stored in a GMP-compliant environment, samples can be reanalyzed, and new information extracted as needed.
EDDI has short turn-around times thanks to its microarray technology. Our validated pipeline revealed no changes in predictive power when using single CpG sites or differentially methylated regions, in contrast with the currently limited reports of methylation assessment in CHO13 for clone selection, which rely on deep sequencing.
The ML models of EDDI have a biological foundation. Methylation sites used for cell density predictions regulate genes active only in highly proliferative cultures15. Moreover, the predictive models for growth and metabolism are interconnected through gene networks confirmed empirically in CHO16.
While not all methylation sites used as predictors can be currently related to that particular parameter, in accordance with the more established human methylation predictive models11, we expect that more targeted experiments will reveal the nuances in this regulation. This connection between methylation changes and process adaptations would allow to take the models one step further and create a Digital Twin, to perform in silico predictions of changes to the bioprocess.
We guess, you have further questions!
Please contact us via this form or book an appointment! We are abolutly ready to discuss all your thoughts!
